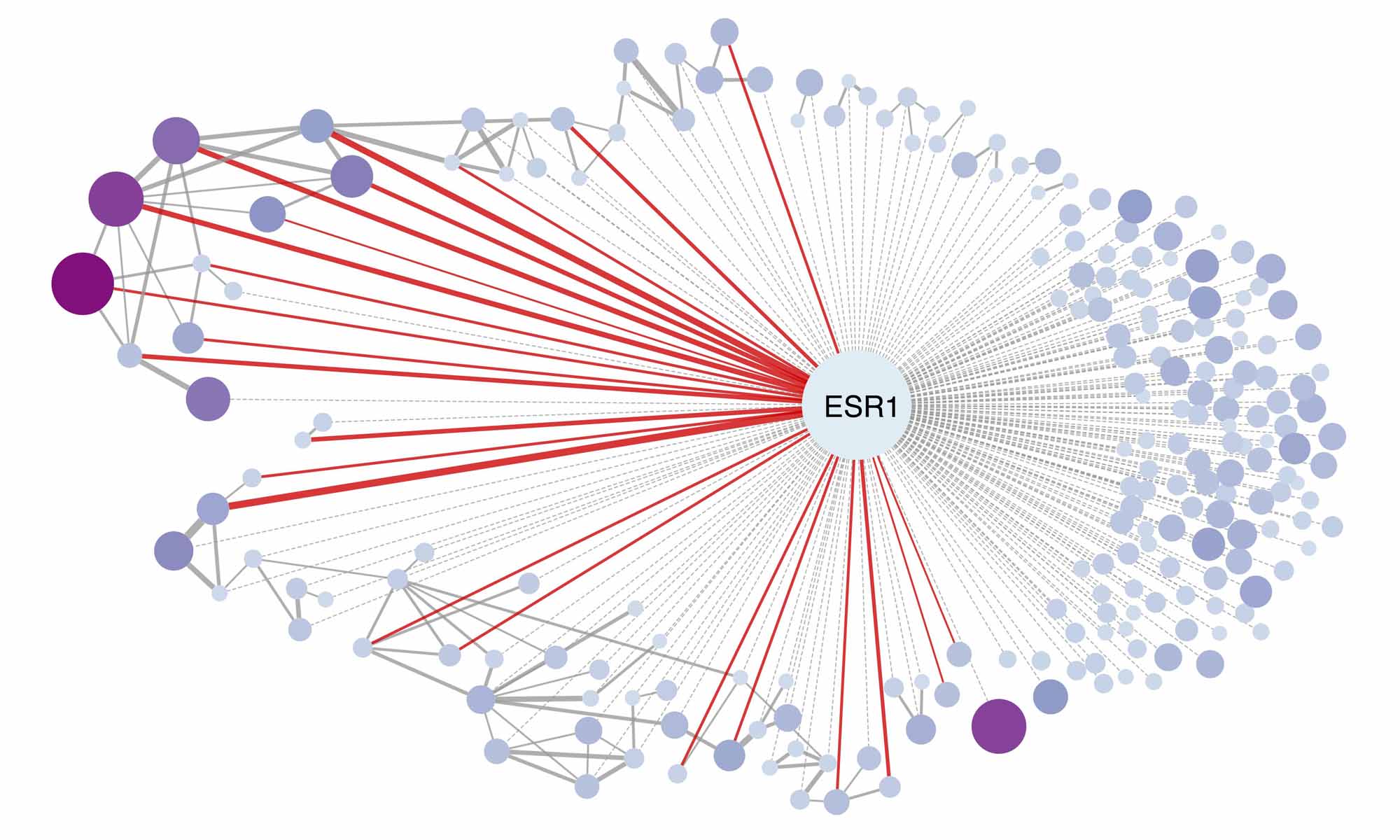
Given that functional similarity exists among genes in the same protein complex or signaling pathway, analysis of coordinated gene effects upon genetic depletion (co-dependency) in large-scale genetic screening may serve as a powerful approach to identify the functional coregulators of NRs. In this regard, we identified the genes whose gene effects were significantly correlated with the gene effects and mRNA expression of a given NR. Additionally, the potential direct interactions between a NR and other genes were also predicted by a computational approach (Dataset 14). As positive controls, many known NR coregulators were successfully identified by our analyses, such as FOXA1 and GATA3 for ESR1; HNF1A and FOXA2 for HNF4A; RARG and KLF5 for RXRA; and certain members in the Mediator complex for PPARG (Dataset 15). Meanwhile, NR-associated genes without direct interaction but coordinate the same cancer-dependent pathway were also observed, for example ESR1 and CDK4 (Figure 6E). Beyond the above known interactions, novel putative molecular interactions were also identified (Dataset 15) and publicly provided to researchers through our Functional Cancer Genome data portal. On average, 87 putative associated genes were predicted for each NR, while 38.3% NRs have predicted direct interaction with the identified NR-associated genes. Taken together, the NR family showed a significantly high ratio of genes defined as strongly selective essential genes in cancer, providing a novel therapeutic opportunity for development of anticancer drugs. The co-dependency and co-expression/dependency information we have collected on NRs also serve as unique resource for further characterizing NR functions in cancer.