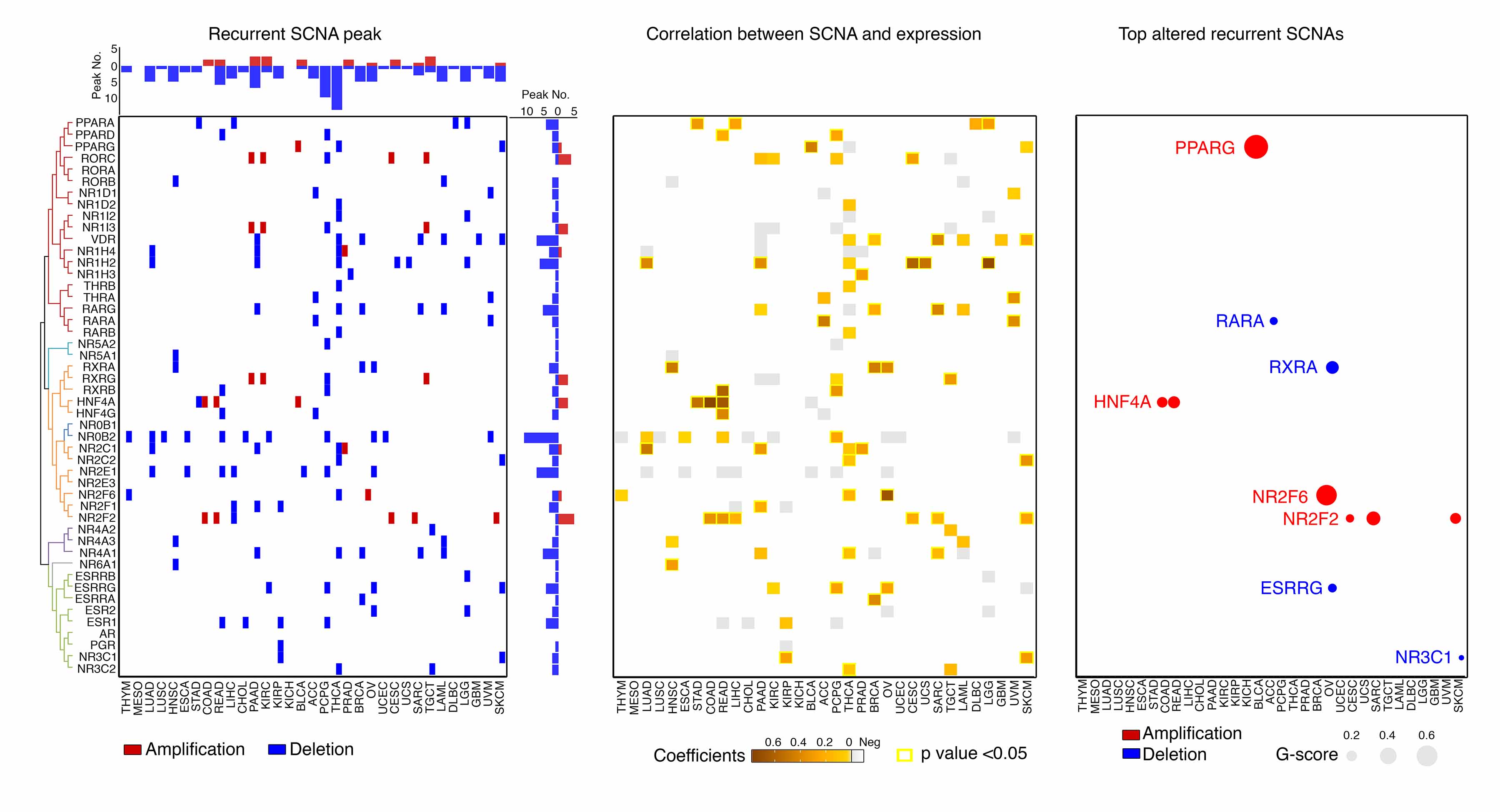
To characterize somatic copy number alterations (SCNAs) of NR genes in cancers, we analyzed single nucleotide polymorphism array profiles from TCGA (Dataset 3). Recurrent focal SCNAs contain only a handful of genes, often exhibit high-amplitude variations ,and are altered more frequently than the background, suggesting the presence of additional selective pressures during tumor cell evolution (i.e., genes in those regions are more likely to play causal roles in tumorigenesis). After estimating the recurrent focal peaks by GISTIC algorithm for each cancer type (q<0.25), NR genes were mapped to these peak regions. Initially, we identified 133 recurrent focal peaks that harbored NR genes across all cancers (Figure 3A and Dataset 7). In 31 of 33 cancer types, excepting mesothelioma (MESO) and kidney chromophobe (KICH), NR genes fell into at least one focal peak. 91.6% (44/48) of NR genes were mapped into at least one recurrent focal peak in one cancer type. Notably, 83.5% (111/133) of the identified NR-harboring peaks were deletion peaks, while only 17% of them were amplification peaks. Next, we analyzed the correlations between mRNA expression levels and copy numbers for the above recurrent focal SCNA events. In 55 of 133 (41.4%) events, which including 27 NR genes in 22 cancer types, significant and positive correlations were observed (Pearson test p-value<0.05; Figure 3B and Dataset 8), indicating that copy number alteration indeed was one of the mechanisms by which NR expression was dysregulated in cancers. Finally, to further identify the most significant NRs driven by recurrent SCNAs, we estimated the G-scores, which considered both the amplitude of the aberration and the frequency of its occurrence across samples, for the above 27 NRs. At a pan-cancer level, PPARG, NR2F6, NR2F2 and HNF4A were the top 4 NRs which showed the highest overall G-scores for copy number gains, while RXRA, ESRRG, RARA and NR3C1 were identified as the 4 most significant NRs with copy number losses (Figure 3C, Dataset 9). Interestingly, for 7 of the above 8 NRs, their recurrent SCNAs were observed in a single cancer type or closely related tissue lineages. Among these NRs, HNF4A and RXRA showed the highest frequencies for copy number gains and losses in given cancer types, respectively (72.4% HNF4A in colon adenocarcinoma [COAD]/rectum adenocarcinoma [READ]; 60.4% RXRA in OV; Figure 3D and 3E). Their mRNA expression levels were significantly and positively correlated with predicted copy numbers in these cancer types (Figure 3F). Notably, although higher frequencies of high-level copy number amplification (GISTIC status=2) were observed, few tumors exhibited high-level copy number deletions (GISTIC status=-2). Supporting this observation, homozygous deletions in NR genes, as estimated with the ABSOLUTE algorithm, were rare genomic events. This result indicates that certain NRs may play essential roles in these cancer types and high-level deletions may be lethal. Taken together, a large percentage of NR genes lost copy numbers during tumorigenesis, which may lead to consequent downregulation of their mRNA expressions. The most significant SCNA-driven NRs were also identified across cancers, including four recurrently and focally amplified NRs that may provide novel targets for cancer treatment.