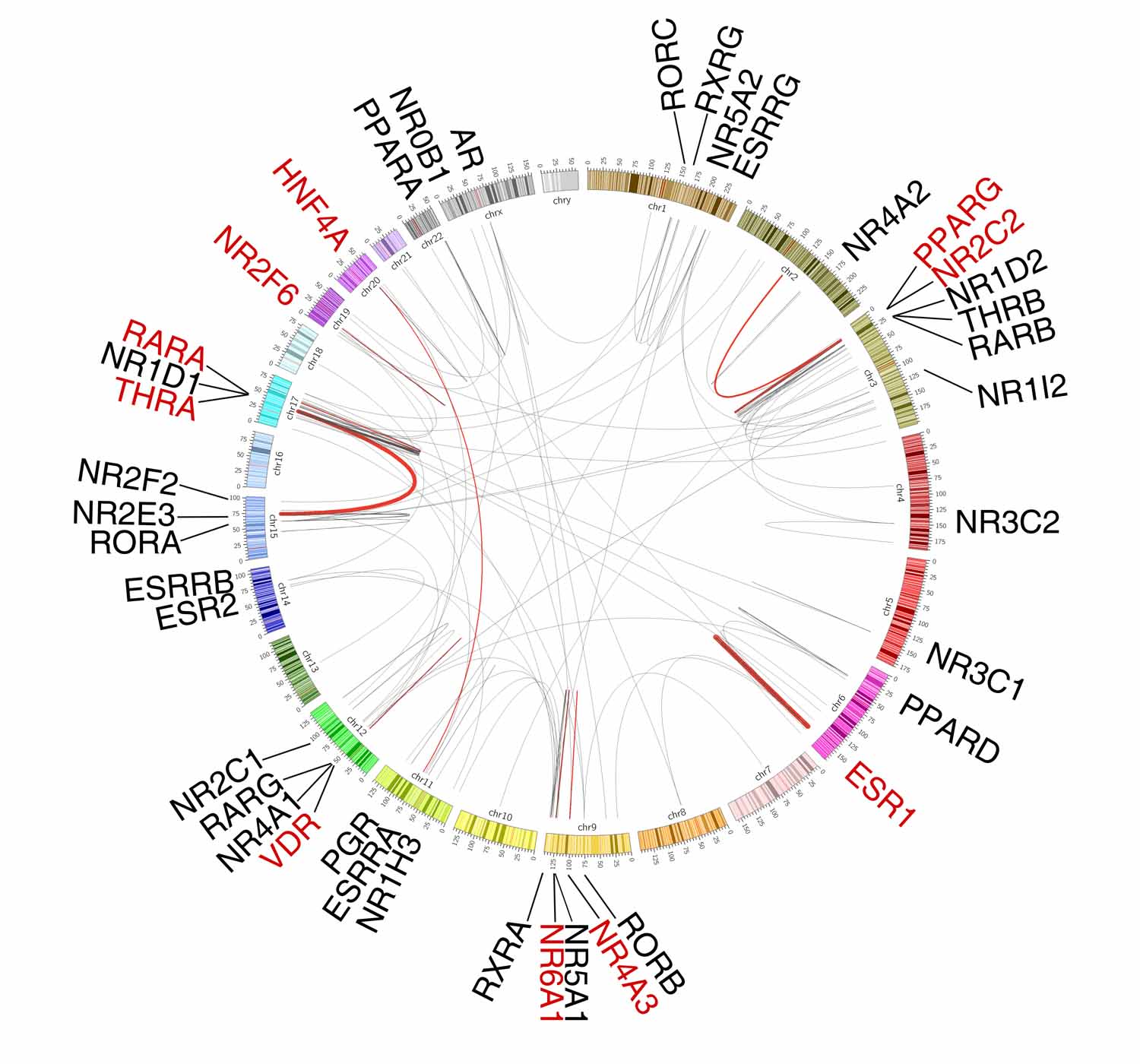
To characterize somatic transcript fusions of NR genes in cancers, we retrieved the processed gene fusion data of TCGA from TumorFusions (Dataset 3). A total of 222 NR fusion events (160 fusion pairs) were identified, including 40 NRs in 28 cancer types (Figure 5A, Dataset 11). More than 90% of these fusion events were classified as Tier 1 and 2 fusions (77% and 14.9%, respectively) by the TumorFusions database. Across cancers, RARA (n=56), ESR1 (n=30) and PPARG (n=17) were the top 3 NR genes that had the highest numbers of fusion events which were largely concentrated in a few cancer types (Figure 5B). 72.9% of the NR genes had less than 5 fusion events across all cancers and no fusion event was found for 8 NR genes (Figure 5C). Acute myeloid leukemia (LAML), UCS and sarcoma (SARC) showed the highest frequencies of NR gene fusions (8.9%, 7.0% and 6.5%, respectively), whereas lymphoid neoplasm diffuse large B-cell lymphoma (DLBC), KICH, pancreatic adenocarcinoma (PAAD), testicular germ cell tumors (TGCT) and uveal melanoma (UVM) had no NR fusion events. 17/160 (10.6%) NR fusion pairs were identified as recurrent fusions (i.e., occurring at least twice across all cancers), including 10 NR genes in 17 cancer types (Figure 5D, Dataset 12). Fusions between PML-RARA (n=26), ESR1-C6orf97 (n=19), and PAX8-PPARG (n=8) were the most frequent recurrent fusion events and showed strong tissue lineage specificity (Figure 5D and 5E). Consistently, oncogenic roles of the above fusions have been reported in acute promyelocytic leukemia, ER positive breast cancer, and thyroid cancer, respectively. Taken together, although transcript fusions of NR genes were relatively rare genomic events across cancers, certain recurrent fusion events were identified and may serve as actionable targets for cancer treatment.