Copy Number
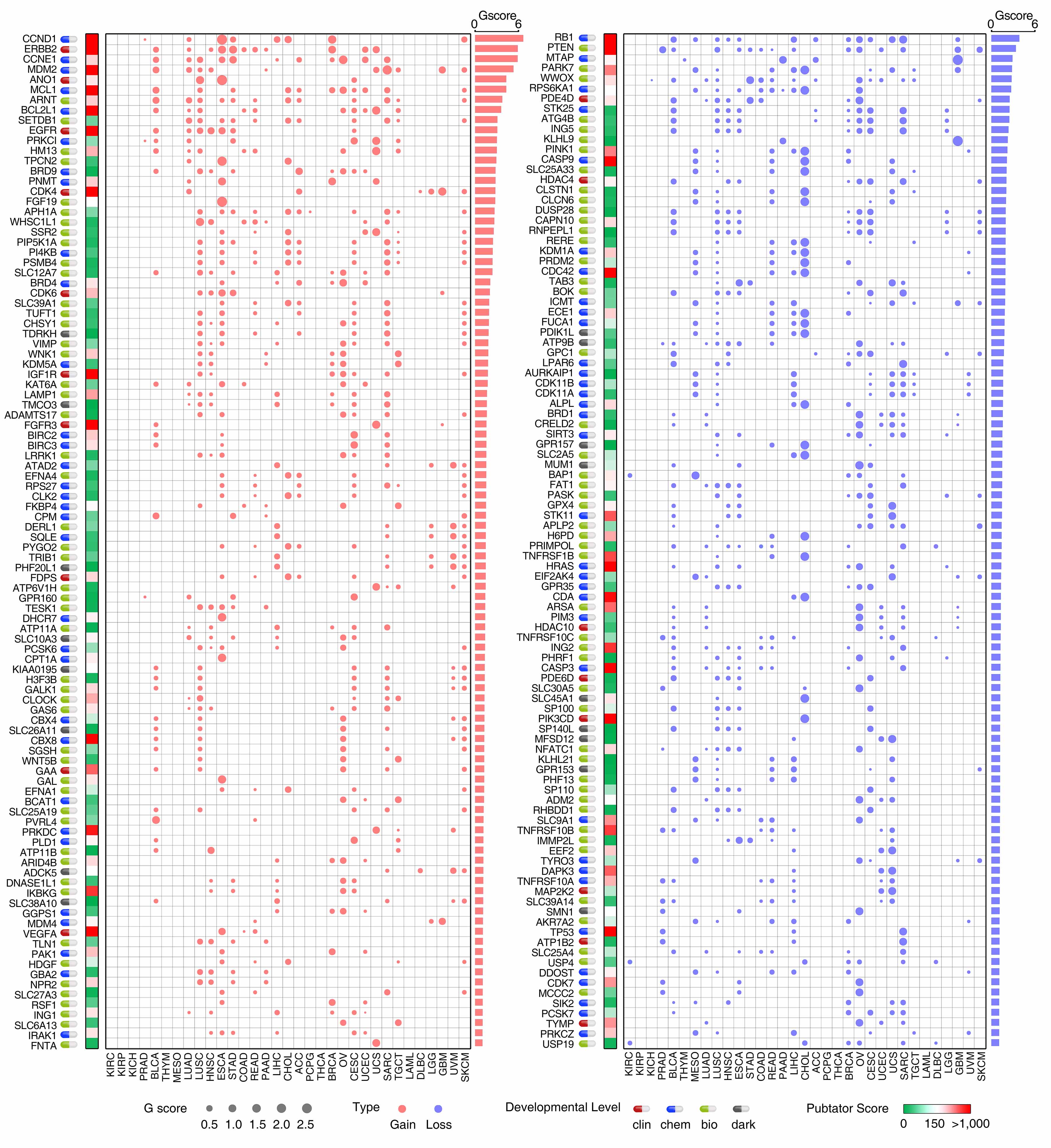
The putative cancer-associated PDGs driven by SCNAs in each cancer type were identified by four criteria: 1) located in a peak region of a significantly recurrent focal SNCA locus estimated by the genomic identification of significant targets in cancer (GISTIC2) algorithm, (q≤0.25); 2) altered with high frequency and large amplitude (G-score ≥0.1); 3) mRNA reliably detected in at least 10% of tumor specimens in the given cancer type (90th percentile of FPKM value ≥1); and 4) mRNA significantly and positively correlated with copy numbers (Pearson P-value less than 0.001).
Summary of the G-score of the PDGs in each cancer type
![]()
List of the putative cancer-causing PDGs driven by SCNAs in each cancer type
![]()