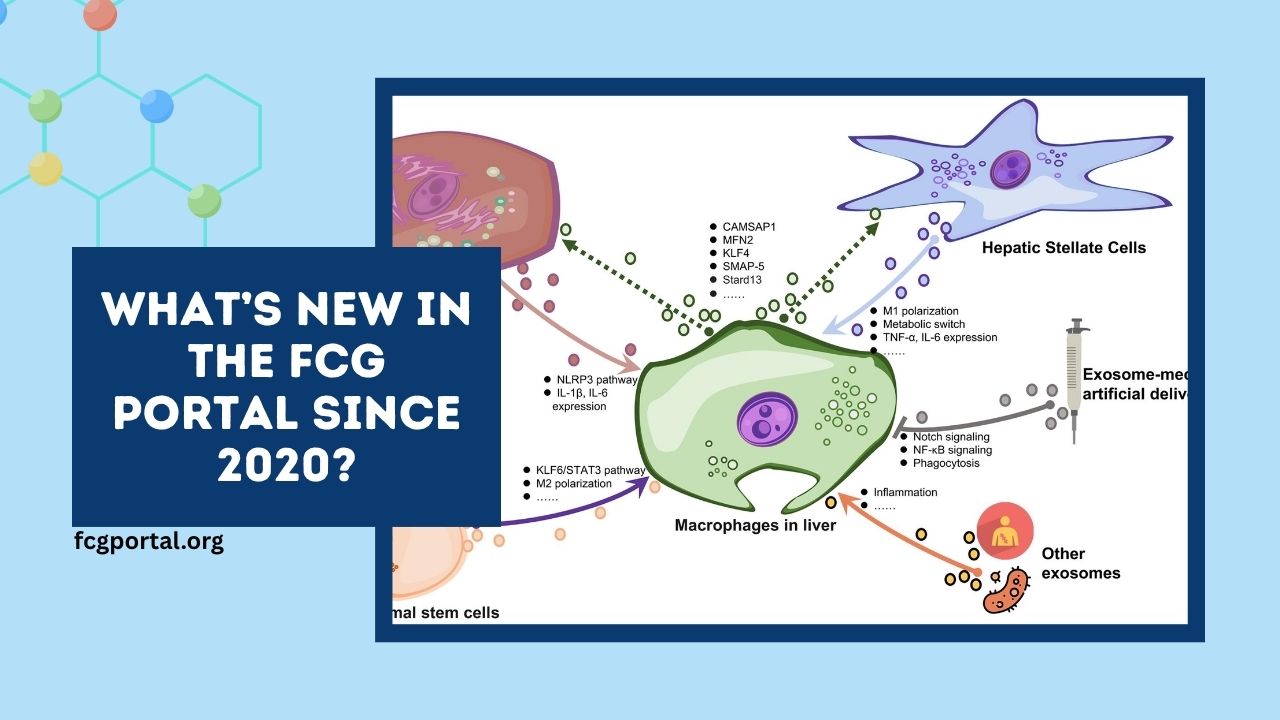
The Functional Cancer Genome (FCG) Portal has evolved significantly since 2020, enriching the cancer research community with new atlases, improved search functionality, streamlined user interface, and refined data accessibility.
These updates underscore FCG’s commitment to advancing cancer genomics tools and maintaining a researcher-friendly platform.
Major FCG Portal Enhancements Since 2020
| Year | Update | Description |
|---|---|---|
| 2020 (March) | TCSA Launched | The Cancer Surfaceome Atlas (TCSA) was introduced to map cell surface proteins across cancers. |
| 2020 (Feb–Mar) | Improved UI and Functionality | URLs structured for better navigation, search function enabled, and page titles optimized for SEO. |
| 2021 | TCSA Code Repository Published | Analysis code for the TCSA (covering receptor-ligand interactions, expression, and growth dependencies) made openly available. |
| Ongoing (since 2018) | Genomic Atlas Expansion | Maintained and expanded key atlases like TCLA, TCGAA, and TCDA, facilitating multi-dimensional cancer-genomic analysis. |
Detailed Summary of Enhancements
1. Launch of the Cancer Surfaceome Atlas (TCSA)
The March 2020 debut of TCSA significantly broadened FCG’s scope, enabling researchers to explore cancer-specific cell surface proteins, their genomic alterations, growth dependencies, and therapeutic potential. This formed a crucial bridge between structural biology and drug-target discovery.
2. Enhanced Usability and Search Optimization
Between February and March 2020, the portal underwent a usability transformation—site URLs were structured, page titles were made SEO-friendly, and a search function was introduced—enabling quicker, more intuitive access to data across atlases like TCCCA and TCGAA.
3. Open-Source Access to Analytical Tools
In 2021, FCG published the TCSA analysis code on GitHub—empowering researchers to dive deeper into datasets encompassing expression profiling, receptor-ligand dysregulation, and identification of 1,278 predicted interactions along with 409 cancer-specific candidate targets.
4. Continued Expansion of Genomic Atlases
Over these years, foundational resources such as the Cancer LncRNome Atlas (TCLA), Cancer Genetic Ancestry Atlas (TCGAA), and Cancer Druggable Gene Atlas (TCDA) remained enriched and updated, reaffirming FCG’s dedication to comprehensive, multi-layered genomic annotation.
Why These Updates Matter
- Improved Research Accessibility: With enhanced search and navigation tools, researchers can quickly locate datasets and insights.
- Accelerated Discovery: The availability of open-source code enhances reproducibility and facilitates novel, customized analyses.
- Expanded Functional Coverage: New atlases like TCSA and ongoing maintenance of TCLA, TCGAA, and TCDA augment FCG’s reach across structural, non-coding, ancestry, and pharmacogenomic domains.
- Stronger Translational Potential: The integration of surfaceome data and druggability annotations equips scientists to pursue biomarkers and therapeutic targets more effectively.
Since 2020, the FCG Portal has undergone substantial upgrades—from launching TCSA to refining search and UI features, and providing open-source analytical tools—solidifying its role as an indispensable resource in cancer genomics and translational oncology.
These enhancements not only improve accessibility and analysis but also empower researchers to accelerate discovery and precision medicine initiatives.
FAQs
What is the significance of the TCSA launched in 2020?
The Cancer Surfaceome Atlas (TCSA) enables identification of cell surface proteins as biomarkers or drug targets, drastically expanding FCG’s functional insights.
How did FCG Portal improve search and navigation?
In early 2020, FCG optimized URLs, implemented SEO-friendly page titles, and introduced a search function, streamlining data access across its atlases.
Why is open-source code for TCSA essential?
Publishing the TCSA analysis code enables transparency, reproducibility, and allows researchers to tailor analyses specific to their research goals.