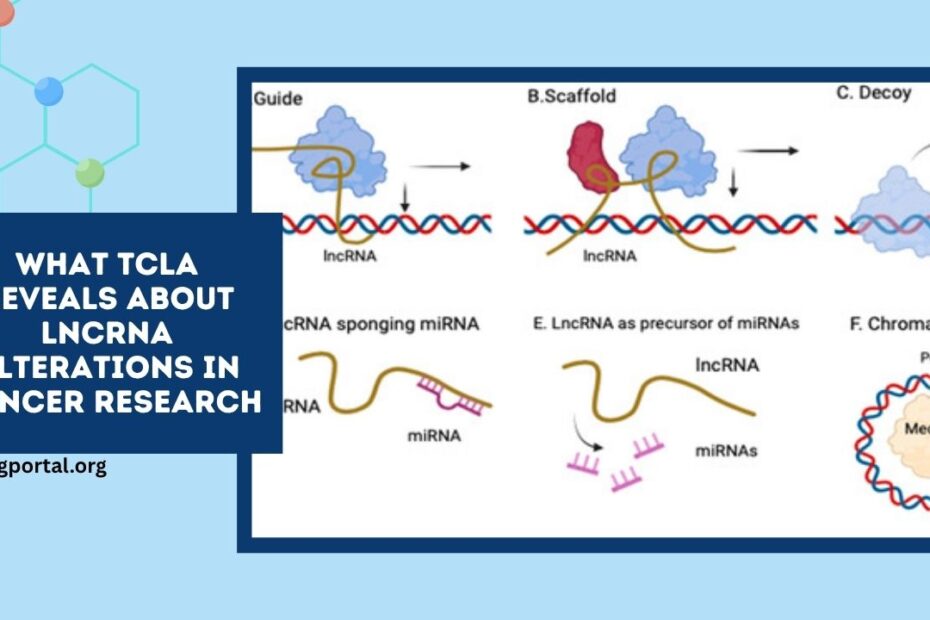
In cancer research, long non‑coding RNAs (lncRNAs) are emerging as critical regulators of tumorigenesis.
The Cancer LncRNome Atlas (TCLA) offers a robust platform to explore how lncRNAs are altered across diverse cancer types—including expression, genomic changes, and epigenetic modifications, powered by large-scale TCGA data.
What Is TCLA?
TCLA is a research database enabling exploration of lncRNA alterations across multiple human cancers.
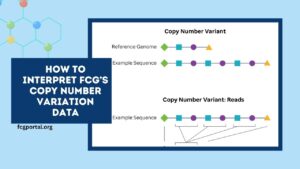
It includes expression, copy‑number variation (CNV), and epigenetic data drawn from TCGA and NCI’s CGHub datasets.
Key Features of TCLA
| Feature | Description |
|---|---|
| Data Sources | TCGA Data Portal & NCI’s Cancer Genomics Hub (CGHub) |
| Number of Cancer Types | 13 distinct cancer types covered |
| Alteration Dimensions | Includes expression, genomic (CNV), and epigenetic data |
| Query Input | Searchable by Ensembl gene accession (e.g. ENSG00000249859) |
Why TCLA Matters
- Pan‑cancer view: It allows researchers to understand how individual lncRNAs behave across different cancer types.
- Multidimensional data: The combination of expression, CNV, and epigenetic alterations offers a comprehensive view of lncRNA behavior—key to unraveling mechanisms of tumor progression.
- Research-ready: Enables hypothesis generation for biomarker discovery, lncRNA functional roles, and therapeutic targeting.
How TCLA Is Used in Research
TCLA empowers scientists to:
- Identify lncRNAs consistently up‑ or down‑regulated in multiple cancers, potentially serving as diagnostic or prognostic biomarkers.
- Correlate CNV hotspots with lncRNA dysregulation that might reflect genomic drivers or vulnerabilities.
- Link epigenetic modifications (e.g., methylation) to expression changes—providing insights into regulatory mechanisms.
Supplementing TCLA, other platforms like TANRIC (an open-access tool covering 20 cancer types) further support lncRNA exploration within a clinical context .
Broader research indicates that lncRNAs regulate translation and contribute to cancer adaptation and plasticity, highlighting their emerging significance beyond transcription.
Meanwhile, lncRNA‑epigenome interactions offer new therapeutic avenues via RNA interference, CRISPR, or small-molecule strategies .
Real-World Insight
While TCLA doesn’t publish specific figures, its foundation on TCGA ensures it draws on thousands of patient samples across 13 cancer types, offering statistically robust insights.
Researchers use TCLA to pinpoint lncRNAs like those with amplification at chromosome 8q24.21, a region frequently altered in many cancers.
TCLA is a pivotal, multi-dimensional lncRNA atlas that empowers cancer researchers to examine expression, genomic, and epigenetic alterations across numerous cancer types.
Leveraging large-scale TCGA data, it enables the discovery of novel lncRNA-driven mechanisms, biomarkers, and therapeutic targets.
In concert with tools like TANRIC and emerging therapeutic strategies targeting lncRNA‑epigenome pathways, TCLA stands at the forefront of precision oncology.
FAQs
What data does TCLA provide about lncRNAs?
TCLA provides transcriptional, copy‑number, and epigenetic alteration data for lncRNAs across a panel of 13 TCGA‑derived cancer types.
How can I search in TCLA?
You can search using an Ensembl gene accession code (such as ENSG00000249859) to explore lncRNA alterations across cancers.
How does TCLA differ from TANRIC?
TCLA focuses on alteration landscapes (expression, CNV, epigenetics) across 13 cancers. TANRIC supports interactive exploration of lncRNA expression in ~20 cancers and links that to clinical and molecular data.