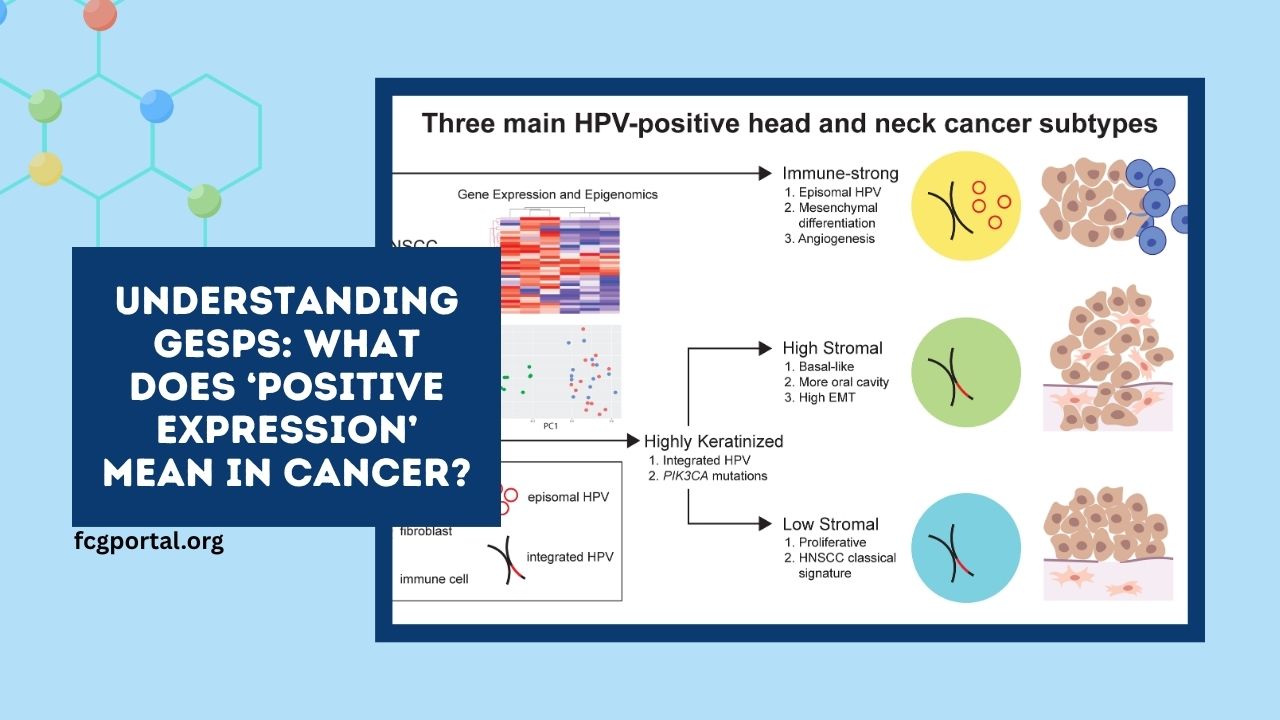
In the evolving landscape of cancer research, identifying and understanding the role of specific proteins on the surface of cancer cells is crucial.
These proteins, known as genes encoding cell surface proteins (GESPs), play a pivotal role in the development and progression of various cancers.
One significant aspect of GESPs is their positive expression, which can influence tumor behavior and therapeutic responses.
What Are GESPs?
Genes encoding cell surface proteins (GESPs) are genes that produce proteins located on the surface of cells. These proteins are involved in various cellular functions, including:
- Cell communication
- Immune response modulation
- Signal transduction
- Cell adhesion and migration
In cancer, the expression levels and alterations of these proteins can significantly impact tumor progression and metastasis.
Defining ‘Positive Expression’ in GESPs
The term positive expression refers to the detectable presence of a particular GESP in a significant proportion of tumor samples.
Specifically, a GESP is considered to have positive expression in a given cancer type if its mRNA expression is reliably detected in at least 50% of tumor specimens.
This is quantified using the fragments per kilobase of transcript per million mapped reads (FPKM) value, with a threshold set at ≥1 for the 50th percentile.
This criterion ensures that the gene’s expression is not only present but also sufficiently robust to be considered biologically relevant.
Importance of Positive Expression in Cancer
The positive expression of GESPs in cancer cells holds several implications:
1. Tumor Classification and Diagnosis
Accurate identification of GESPs with positive expression can aid in classifying tumors more precisely, leading to better diagnostic accuracy and personalized treatment strategies.
2. Prognostic Indicators
Certain GESPs with positive expression are associated with aggressive tumor behavior and poor prognosis. Monitoring these markers can help in assessing disease progression and potential outcomes.
3. Therapeutic Targeting
Many FDA-approved anticancer therapies target GESPs. For instance, approximately 65% of these therapies are designed to interact with cell surface proteins, highlighting the therapeutic potential of GESPs with positive expression.
Overview of GESPs in Cancer
| Aspect | Details |
|---|---|
| Definition | Genes producing proteins located on the cell surface involved in various cellular functions. |
| Positive Expression | Detected in ≥50% of tumor specimens with an FPKM ≥1. |
| Role in Cancer | Influence tumor growth, metastasis, and response to therapies. |
| Therapeutic Relevance | Targeted by approximately 65% of FDA-approved anticancer therapies. |
| Prognostic Value | Associated with tumor aggressiveness and patient outcomes. |
Mechanisms Influencing GESP Expression
Several factors can influence the expression levels of GESPs in cancer:
- Genomic Alterations: Mutations, amplifications, or deletions in genes can lead to altered expression levels.
- Epigenetic Modifications: Changes in DNA methylation and histone modifications can regulate gene expression.
- Transcriptional Regulation: Variations in transcription factor activity can affect GESP expression.
- Post-Transcriptional Modifications: RNA splicing and stability can influence the abundance of GESP mRNA.
Understanding these mechanisms is crucial for developing strategies to modulate GESP expression for therapeutic purposes.
Clinical Implications of GESP Positive Expression
1. Personalized Therapy
Identifying GESPs with positive expression can guide clinicians in selecting appropriate therapies. For example, tumors expressing certain GESPs may respond better to specific monoclonal antibodies or immune checkpoint inhibitors.
2. Monitoring Disease Progression
Regular assessment of GESP expression levels can serve as biomarkers for monitoring disease progression or recurrence, allowing for timely intervention.
3. Developing New Therapeutic Targets
Understanding the role of GESPs in cancer can lead to the development of novel therapeutic agents targeting these proteins, offering new avenues for treatment.
Challenges and Future Directions
While the study of GESPs has provided valuable insights into cancer biology, several challenges remain:
- Heterogeneity of GESP Expression: Variability in GESP expression across different tumor types and even within the same tumor can complicate therapeutic targeting.
- Technical Limitations: Current detection methods may not be sensitive enough to detect low-abundance GESPs.
- Functional Validation: Further research is needed to fully understand the functional roles of GESPs in cancer progression.
Future research focused on overcoming these challenges will enhance our ability to utilize GESPs in cancer diagnosis and therapy effectively.
FAQs
What does ‘positive expression’ of GESPs mean in cancer?
‘Positive expression’ indicates that a specific GESP is detectable in at least 50% of tumor samples, with a minimum mRNA expression level quantified by an FPKM ≥1.
How does GESP positive expression affect cancer treatment?
The positive expression of certain GESPs can serve as biomarkers for selecting targeted therapies and monitoring disease progression.
Are all GESPs with positive expression relevant for cancer therapy?
Not all GESPs with positive expression are suitable targets for therapy. Their relevance depends on their role in tumor biology and their potential as therapeutic targets.
Conclusion
Understanding the concept of positive expression in genes encoding cell surface proteins (GESPs) is pivotal in cancer research and therapy. It provides insights into tumor biology, aids in the development of targeted therapies, and helps in monitoring disease progression. As research advances, the role of GESPs in cancer will continue to be a focal point, offering hope for more effective and personalized treatment strategies.