Sub URLs (TCCCA) added.
|
|

|
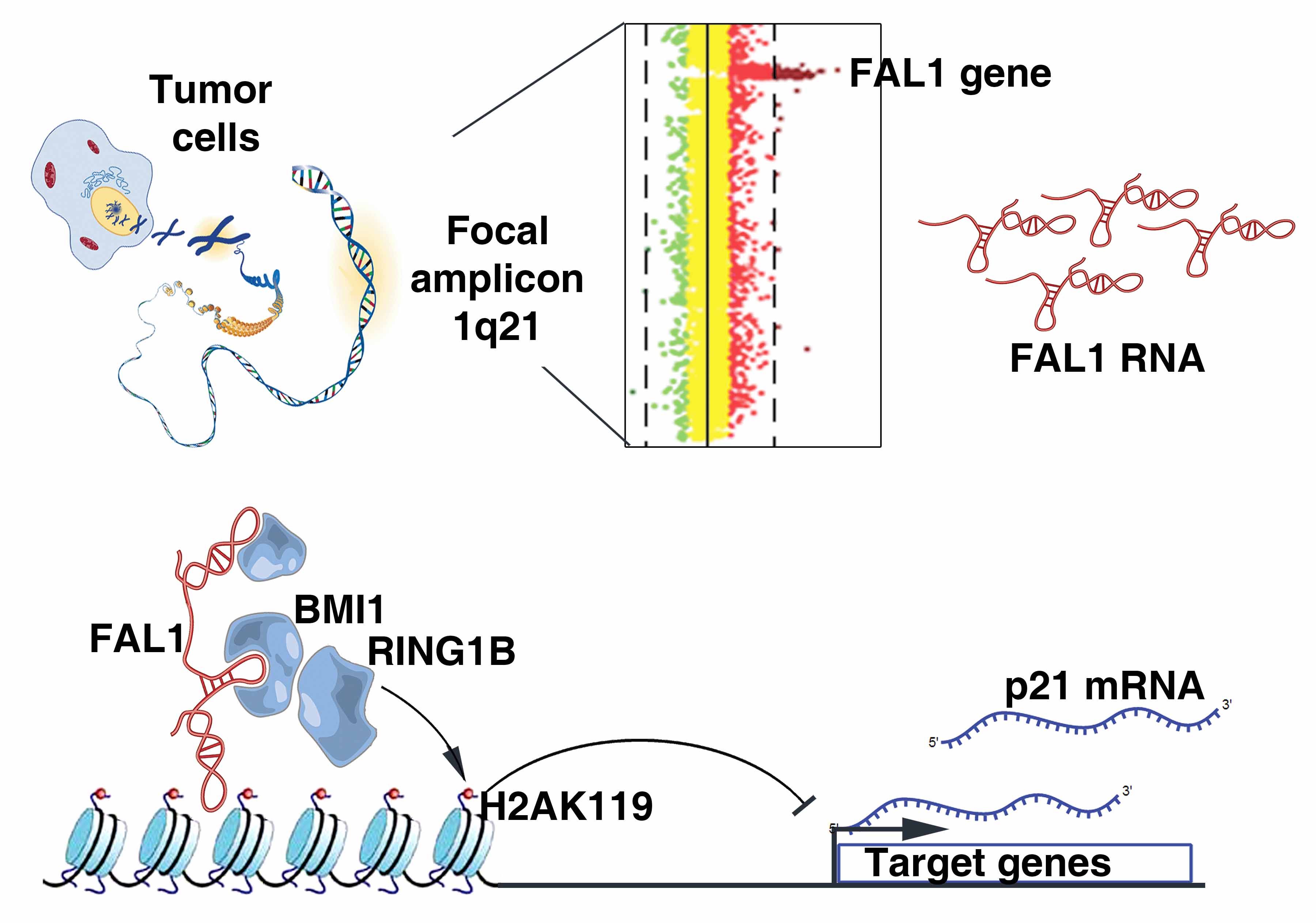
|
Cancer Cell. 2014; 26(3):344-57.
Hu X, Feng Y, Zhang D, Zhao SD, Hu Z, Greshock J, Zhang Y, Yang L, Zhong X, Wang L-P, Jean S, Li C, Huang Q, Katsaros D, Montone KT, Tanyi JL, Lu Y, Boyd J, Nathanson KL, Li H, Mills GB, Zhang L. A Functional Genomic Approach Identifies FAL1 as an Oncogenic Long Noncoding RNA that Associates with BMI1 and Represses p21 Expression in Cancer.
|
||||||||||
|
Cancer Cell. 2015; 28(4):529-40.
Yan X, Hu Z, Feng Y, Hu X, Yuan J, Zhao SD, Zhang Y, Yang L, Shan W, He Q, Fan L, Kandalaft LE, Tanyi JL, Li C, Yuan CX, Zhang D, Yuan H, Hua K, Lu Y, Katsaros D, Huang Q, Montone K, Fan Y, Coukos G, Boyd J, Sood AK, Rebbeck T, Mills GB, Dang CV, Zhang L. Comprehensive Genomic Characterization of Long Non-coding RNAs across Human Cancers.
|
||||||||||
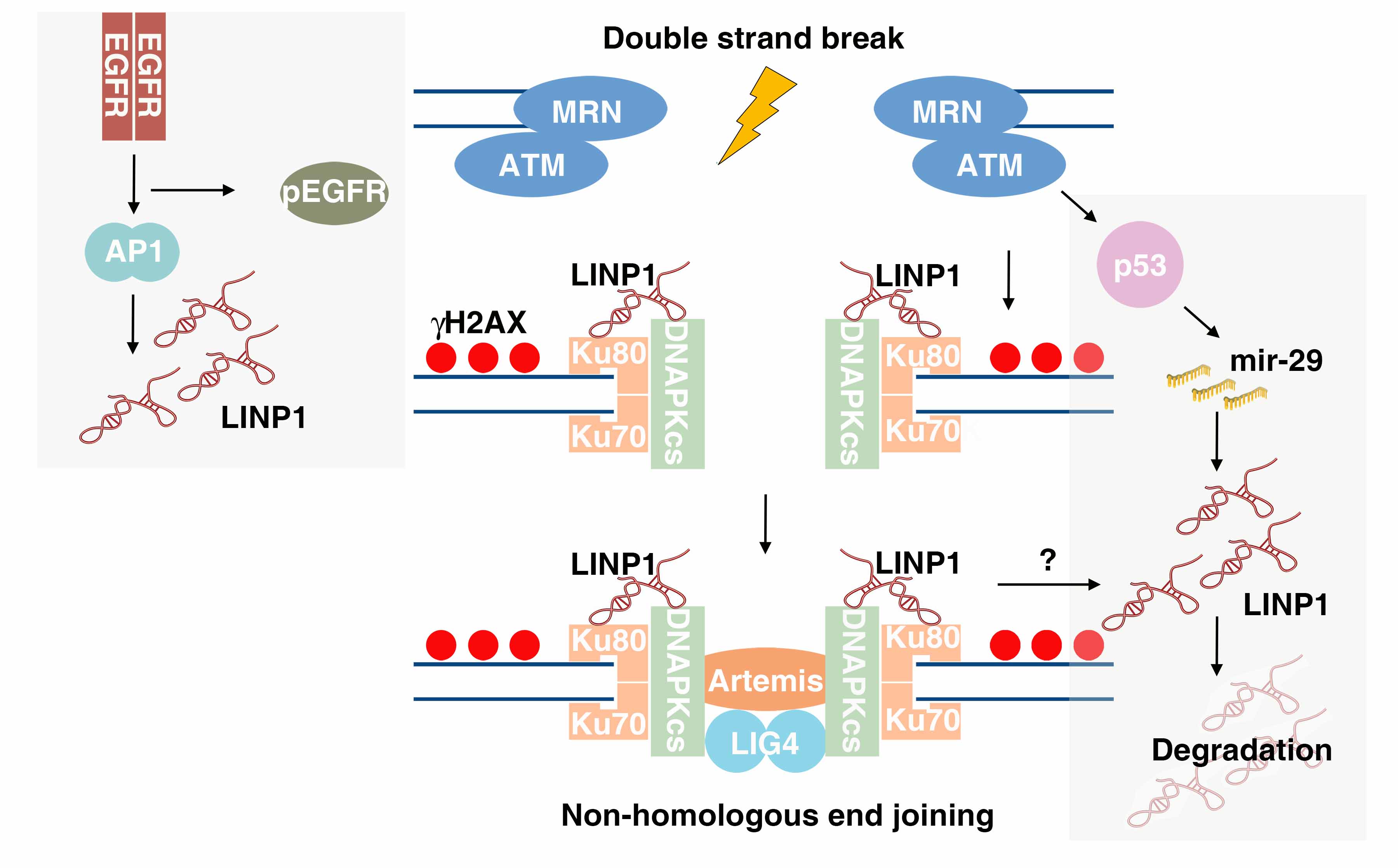
|
Nature Structural & Molecular Biology. 2016; 23(6):522-30.
Zhang Y, He Q, Hu Z, Feng Y, Fan L, Tang Z, Yuan J, Shan W, Li C, Hu X, Tanyi JL, Fan Y, Huang Q, Montone K, Dang CV, Zhang L. Long noncoding RNA LINP1 regulates repair of DNA double-strand breaks in triple-negative breast cancer.
|
||||||||||
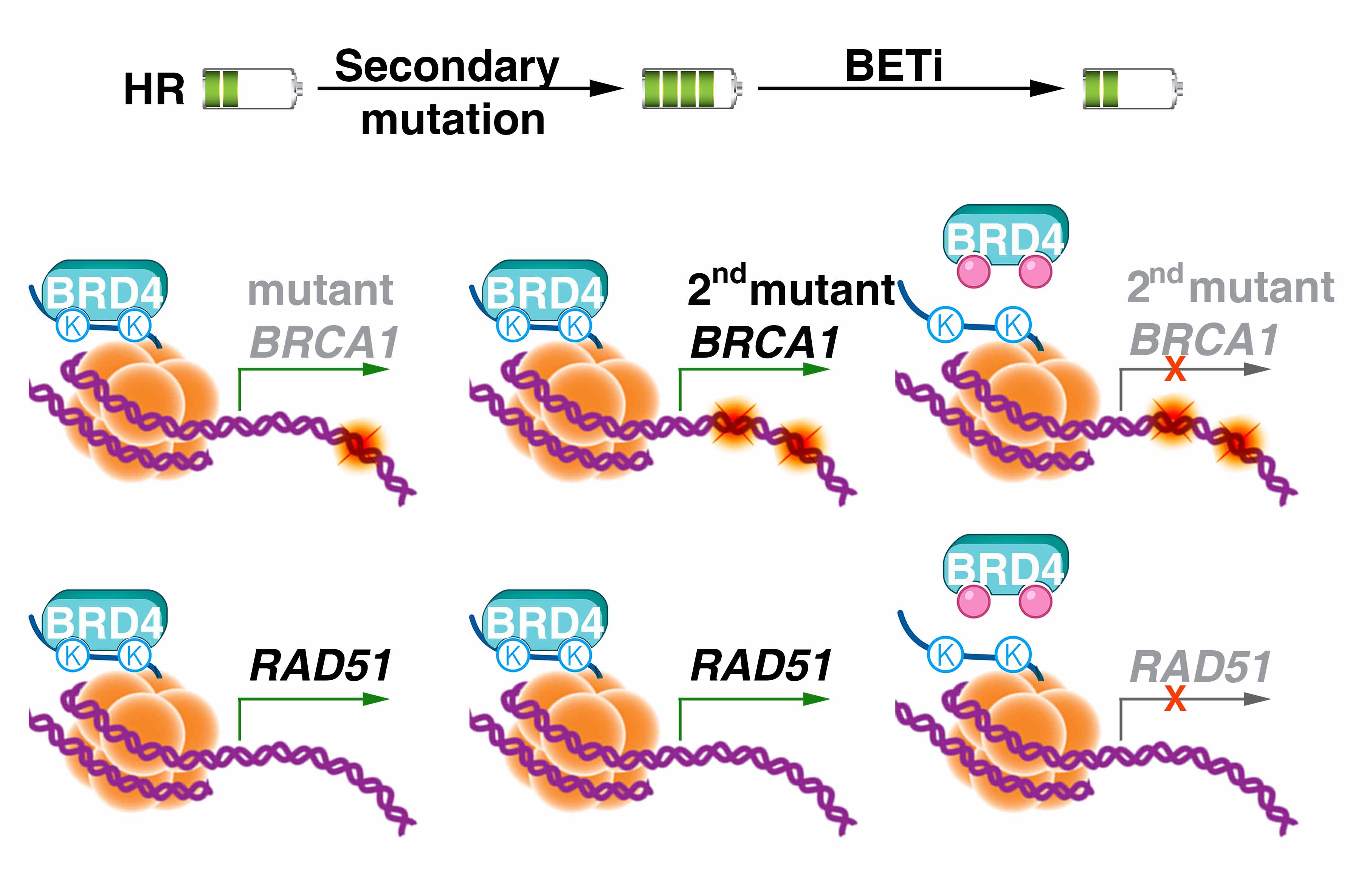
|
Science Translational Medicine. 9, 2017.
Zhang Y, Yang L, Zhang Y, Shan W, Hu Z, Yuan J, Pi J, Wang Y, Fan L, Tang Z, Li C, Hu X, Tanyi JL, Fan Y, Huang Q, Montone K, Dang CV, Zhang L. Repression of BET activity sensitizes homologous recombination-proficient cancers to PARP inhibition.
|
||||||||||
|
Cancer Cell. 2018;34(4):549-60
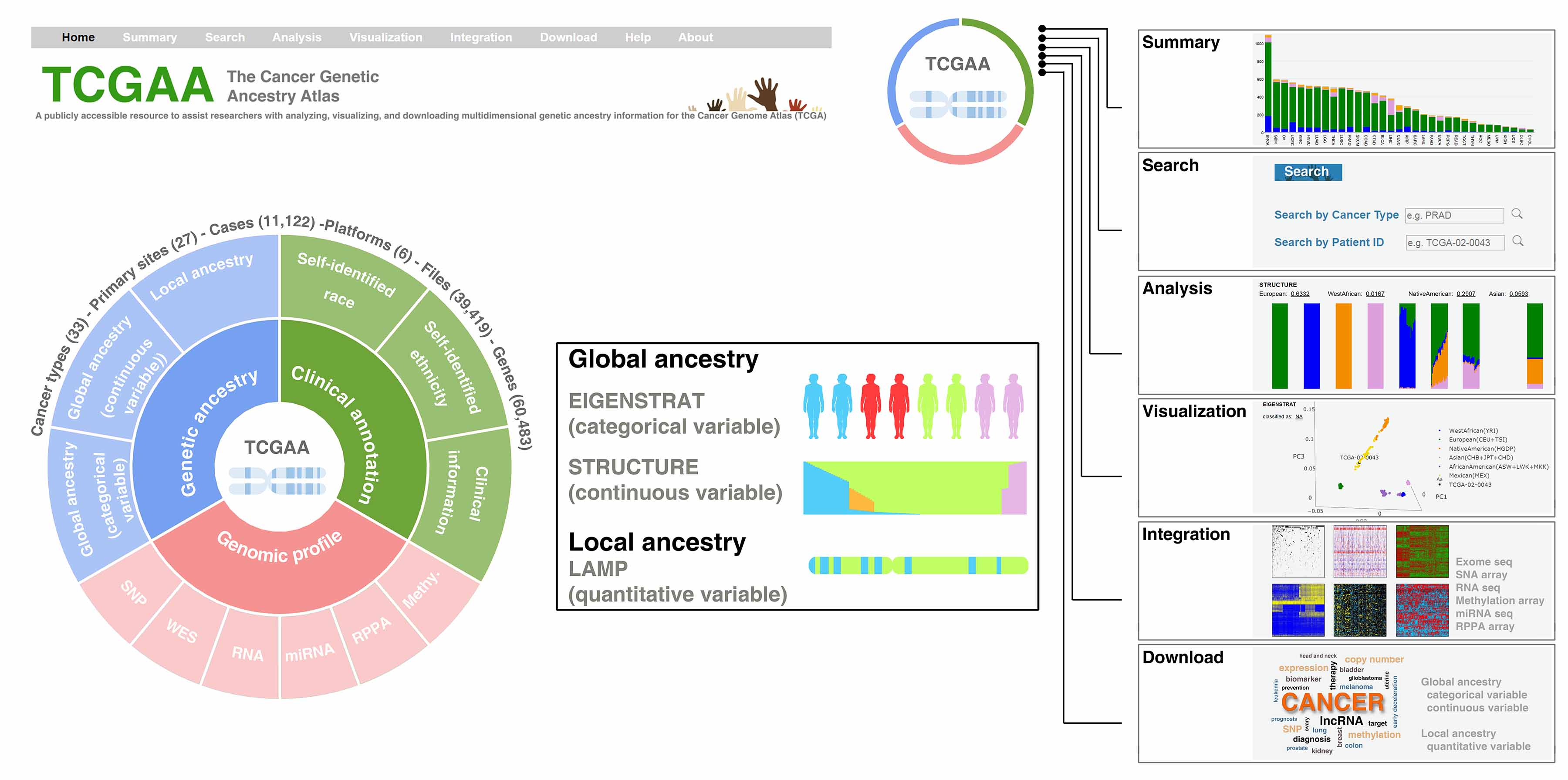
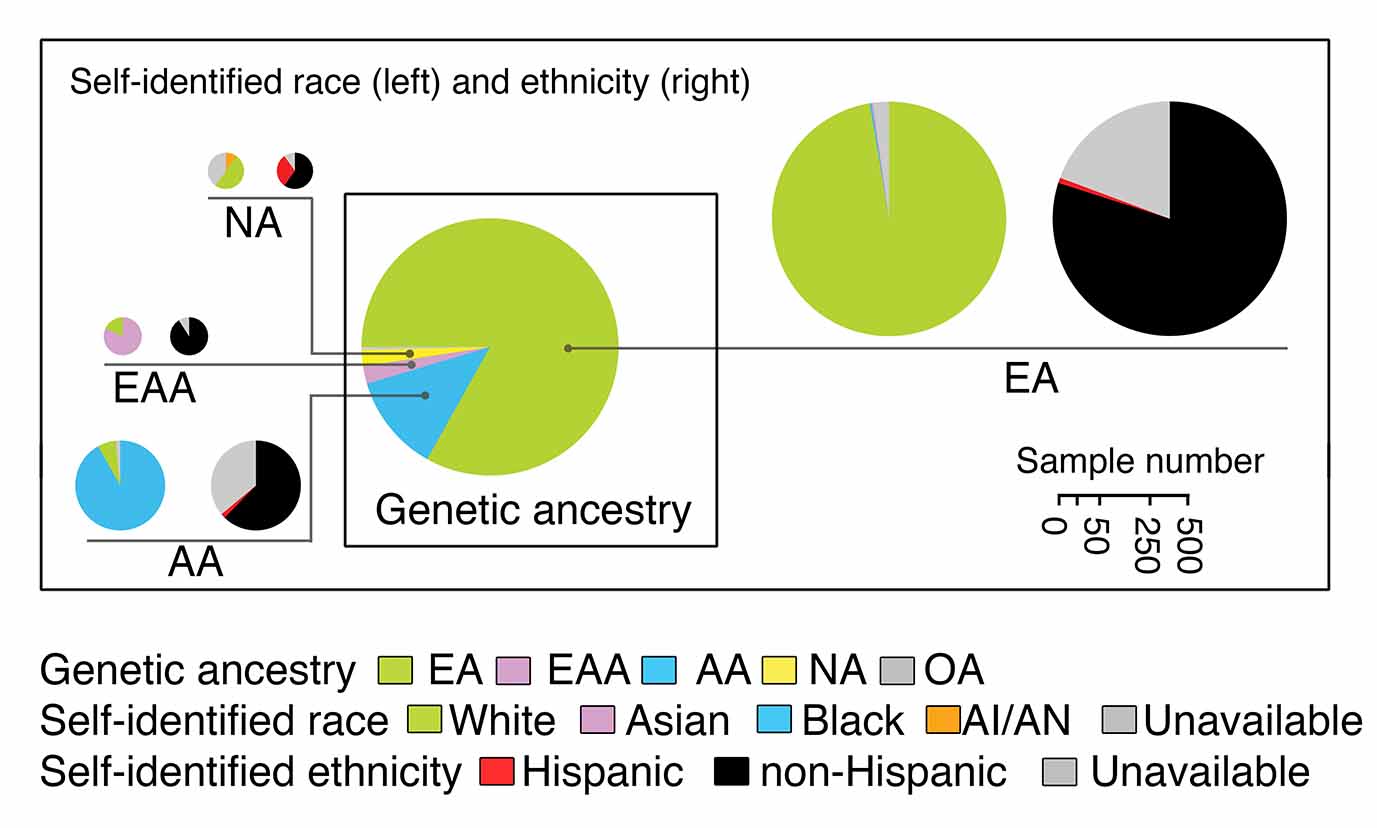
Yuan J, Hu Z, Mahal B, Zhao S, Kensler K, Pi J, Hu X, Wang Y, Jiang J, Zhang Y, Li C, Zhong X, Montone K, Guan G, Tanyi J, Fan Y, Xu X, Long M, Morgan M, Zhang Y, Zhang R, Sood A, Rebbeck T, Dang C, Zhang L. Integrated analysis of genetic ancestry and genomic alterations across cancers.
|
||||||||||
|
Nature Communications. 10(1): 733 (2019)
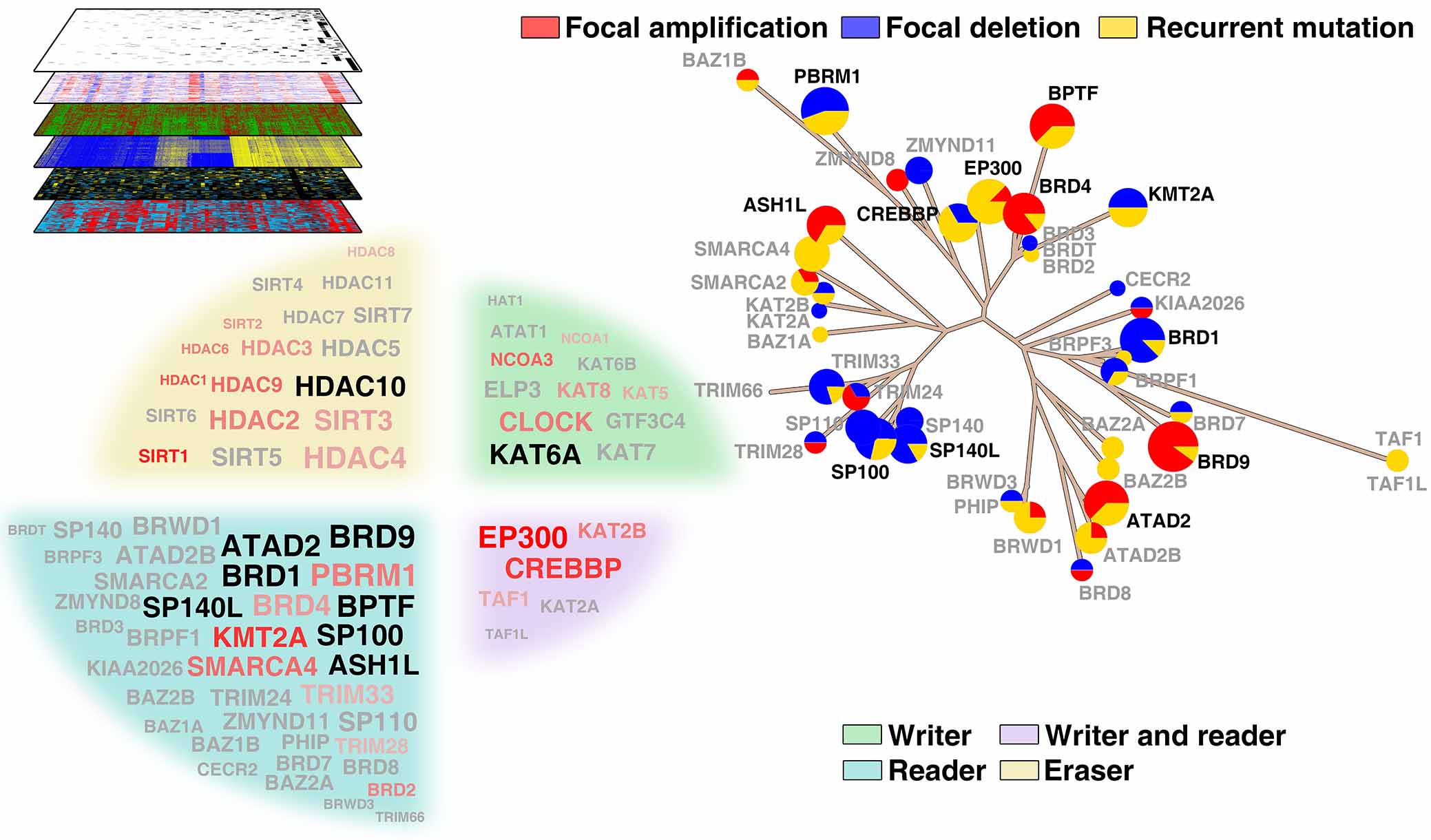
Hu Z, Zhou J, Jiang J, Yuan J, Zhang Y, Wei X, Loo N, Wang Y, Pan Y, Zhang T, Zhong X, Long M, Montone K, T, Tanyi J, L, Fan Y, Wang T, L, Shih I, M, Hu X, Zhang L. Genomic characterization of genes encoding histone acetylation modulator proteins identifies therapeutic targets for cancer treatment.
|
||||||||||
|
PLoS Genet. 2020 Feb; 16(2): e1008641
Yuan, J., Kensler, K. H., Hu, Z., Zhang, Y., Zhang, T., Jiang, J., Xu, M., Pan, Y., Long, M., Montone, K. T., Tanyi, J. L., Fan, Y., Zhang, R., Hu, X., Rebbeck, T. R., Zhang, L. Integrative comparison of the genomic and transcriptomic landscape between prostate cancer patients of predominantly African or European genetic ancestry
|
||||||||||
|
Cell Rep. 2020 Jul 14;32(2):107884
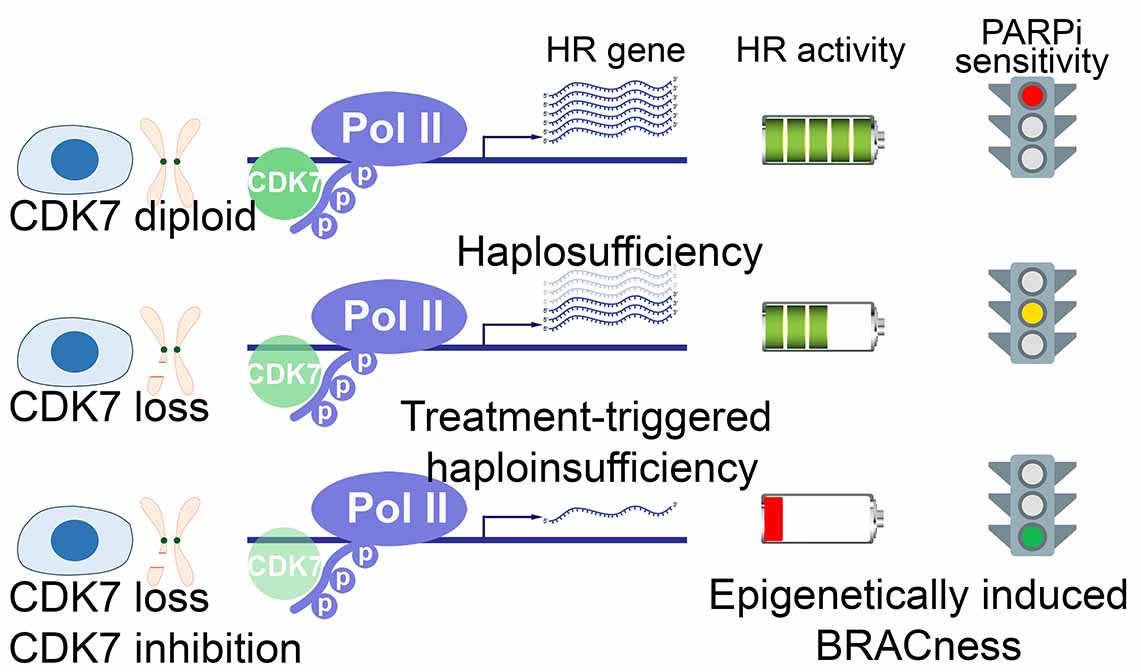
Shan, W., Yuan, J., Hu, Z., Jiang, J., Wang, Y., Loo, N., Fan, L., Tang, Z., Zhang, T., Xu, M., Pan, Y., Lu, J., Long, M., Tanyi, J. L., Montone, K. T., Fan, Y., Hu, X., Zhang, Y., Zhang, L. Systematic Characterization of Recurrent Genomic Alterations in Cyclin-Dependent Kinases Reveals Potential Therapeutic Strategies for Cancer Treatment
|
||||||||||
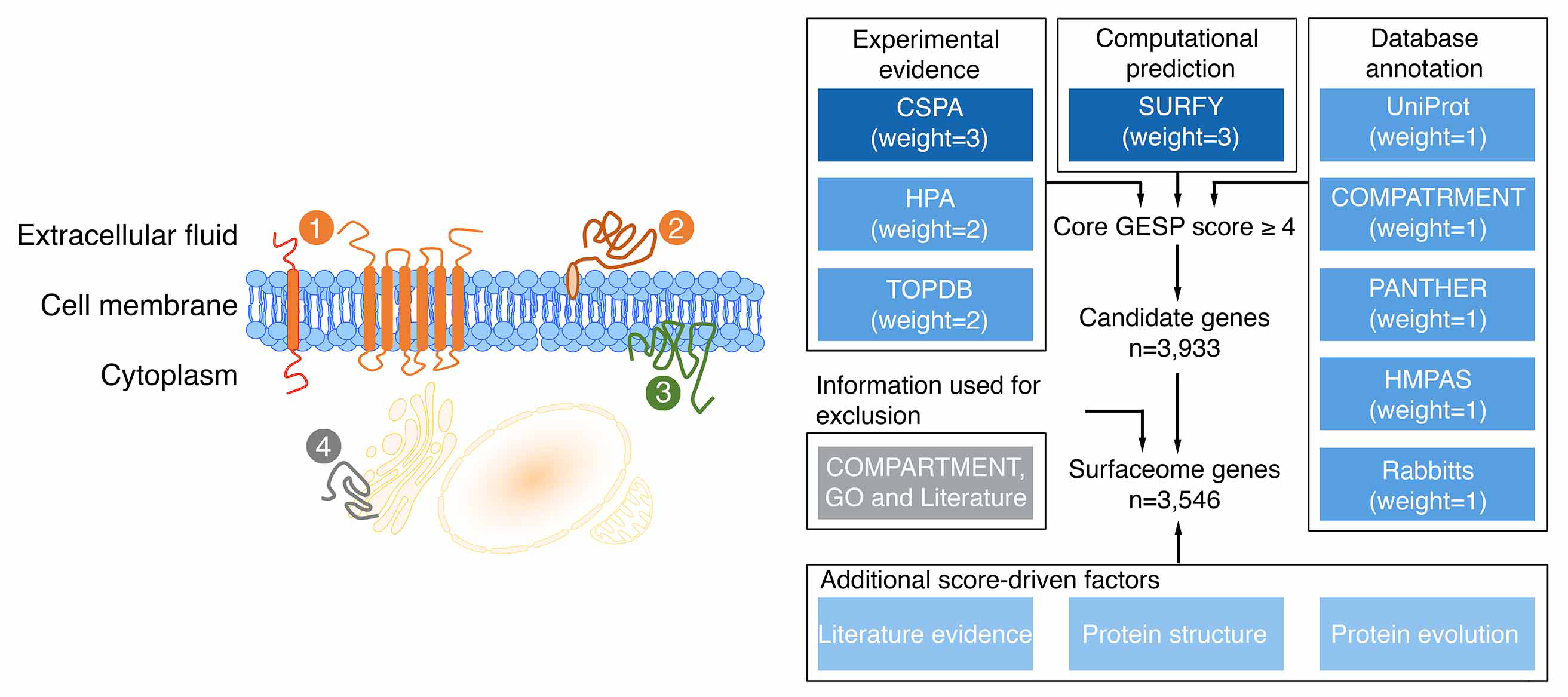
|
Nature Cancer. 2021; 2(12):1406-1422.
Hu Z, Yuan J, Long M, Jiang J, Zhang Y, Zhang T, Xu M, Fan Y, Tanyi JL, Montone KT, Tavana O, Chan HM, Hu X, Vonderheide RH, Zhang L. The Cancer Surfaceome Atlas integrates genomic, functional and drug response data to identify actionable targets.
|
||||||||||
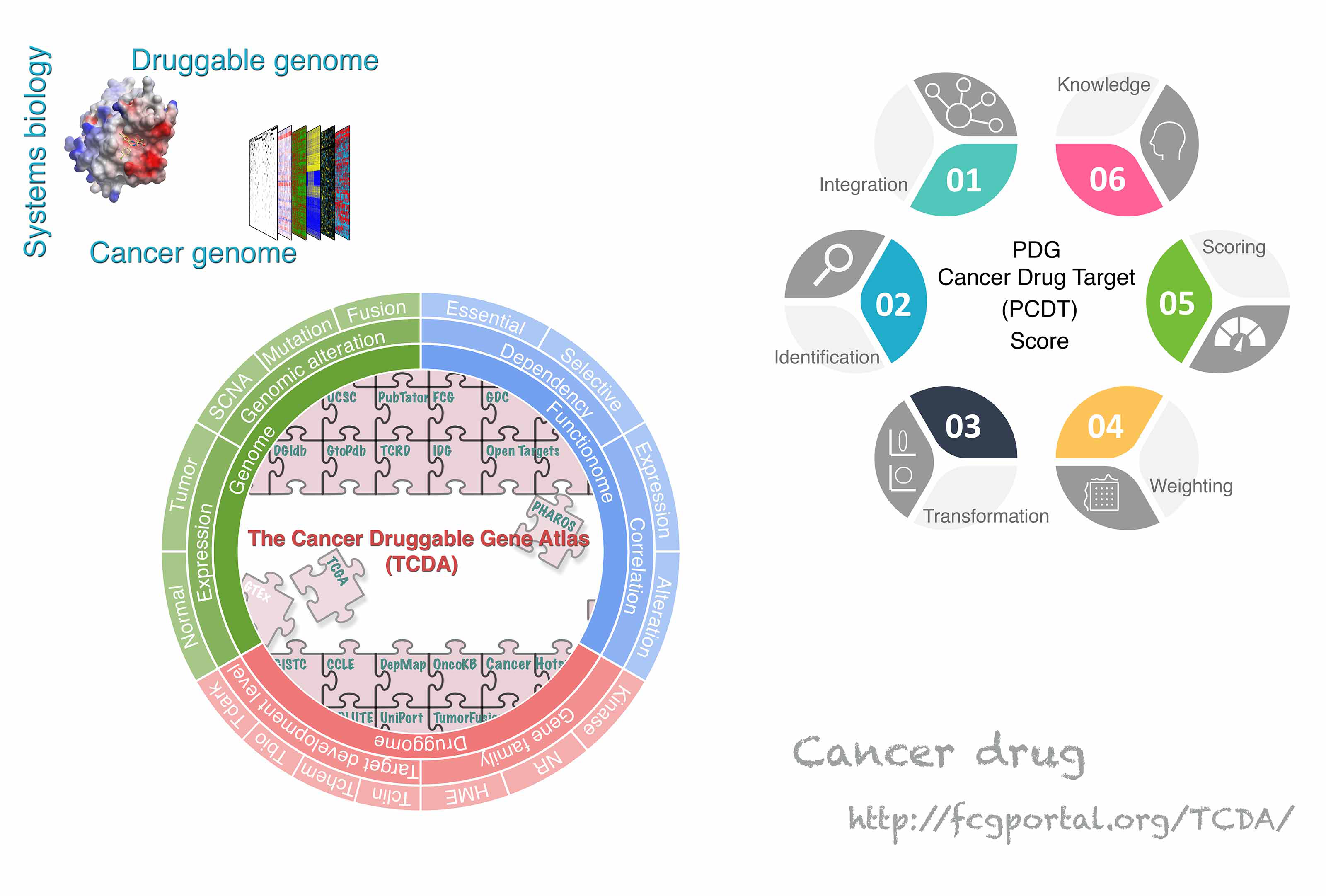
|
Cell Rep. 2022; 22;38(8):110400.
Jiang J, Yuan J, Hu Z, Zhang Y, Zhang T, Xu M, Long M, Fan Y, Tanyi JL, Montone KT, Tavana O, Vonderheide RH, Chan HM, Hu X, Zhang L. Systematic illumination of druggable genes in cancer genomes.
|
||||||||||
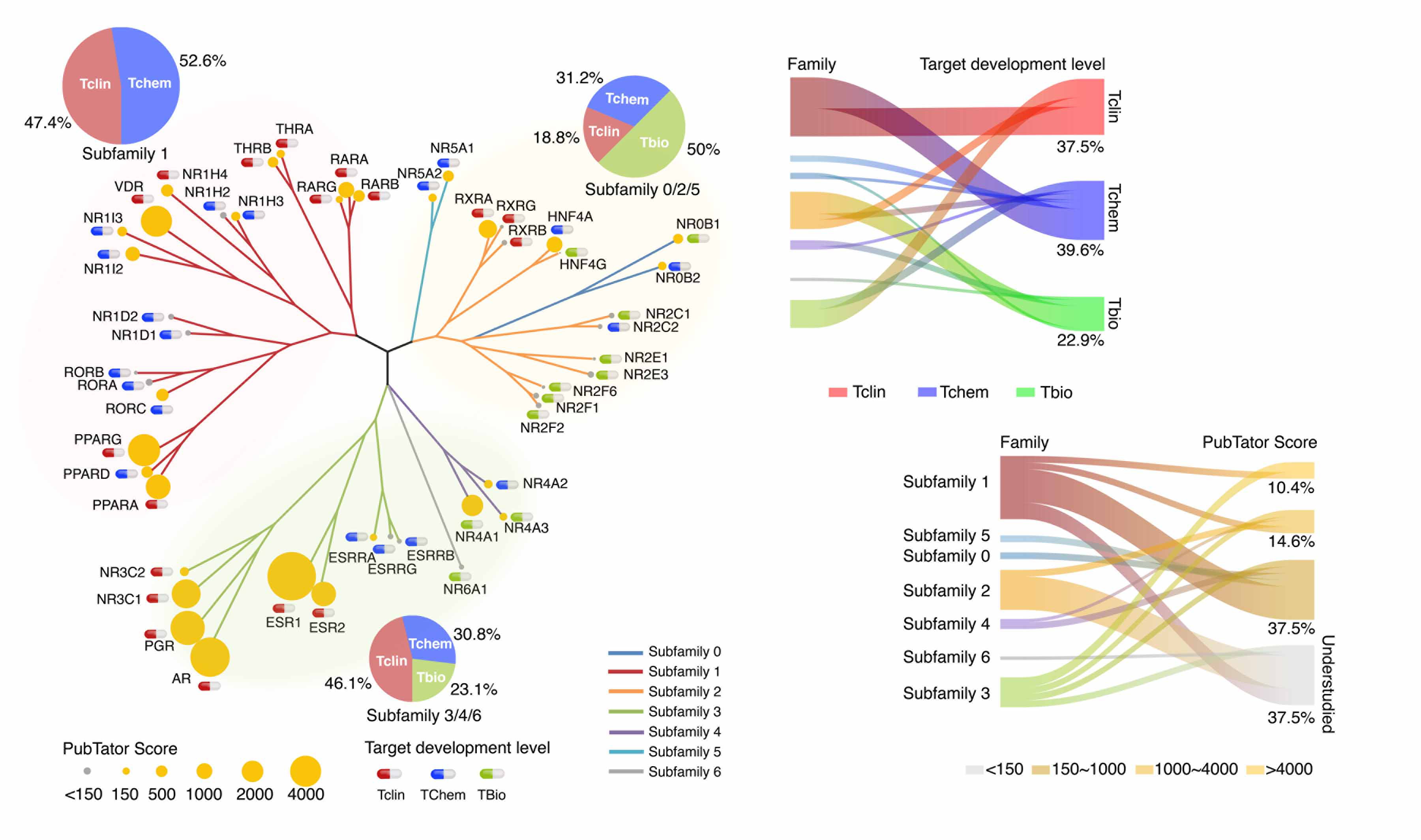
|
Cancer Research. 2022 1;82(1):46-59.
Jiang J, Yuan J, Hu Z, Xu M, Zhang Y, Long M, Fan Y, Montone KT, Tanyi JL, Tavana O, Chan HM, Zhang L, Hu X. Systematic Pan-Cancer Characterization of Nuclear Receptors Identifies Potential Cancer Biomarkers and Therapeutic Targets.
|
||||||||||